| | | | | |
| | | | | |
| |
Home |
Search |
Help |
Download |
|
In the home page there is the possibility to sumbit a quick search using as keyword the name of the microRNA (e.g. 'hsa-mir-211' or 'mir-211'),
writing a list of microRNAs in a text area (example).
or uploading a file with a list of microRNA in the appropriate format with a list of microRNA.
|
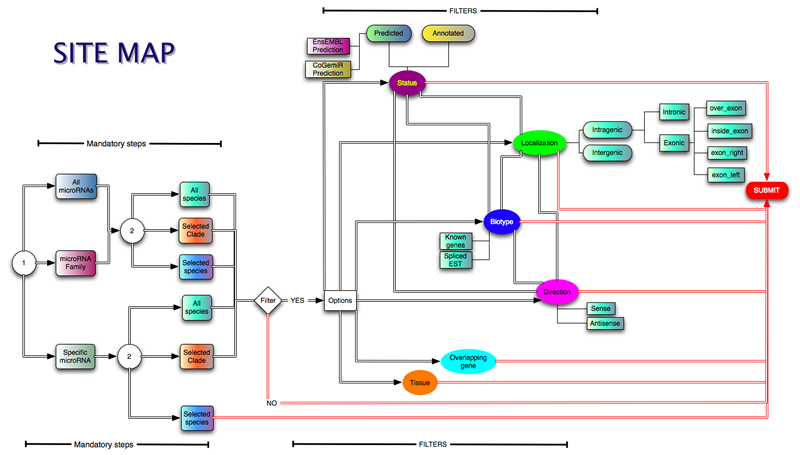
The query mask allows the user to perform more complex queries. In the advanced search all the options are available.
In the main search page the query became more and more specific, depending on the user's choice.
|
|